Quantitative Bio-Image Analysis using Python
Contents
Quantitative Bio-Image Analysis using Python#
This Jupyter book contains training resources for scientists who want to dive into image processing with Python. It specifically aims for students and scientists working with microscopy images in the life sciences. We presume the attendees have some basic programming and image analysis knowledge. To get everyone on the same level, we start with Python programming basics, and image analysis basics, we then dive into descriptive statistics for working with measurements and matplotlib and seaborn for plotting results. We will process images using numpy, scipy, scikit-image and clEsperanto. We will explore Napari for interactive image data analysis. Finally, we will use scikit-learn and StarDist to process images using machine learning and deep learning techniques.
Timetable#
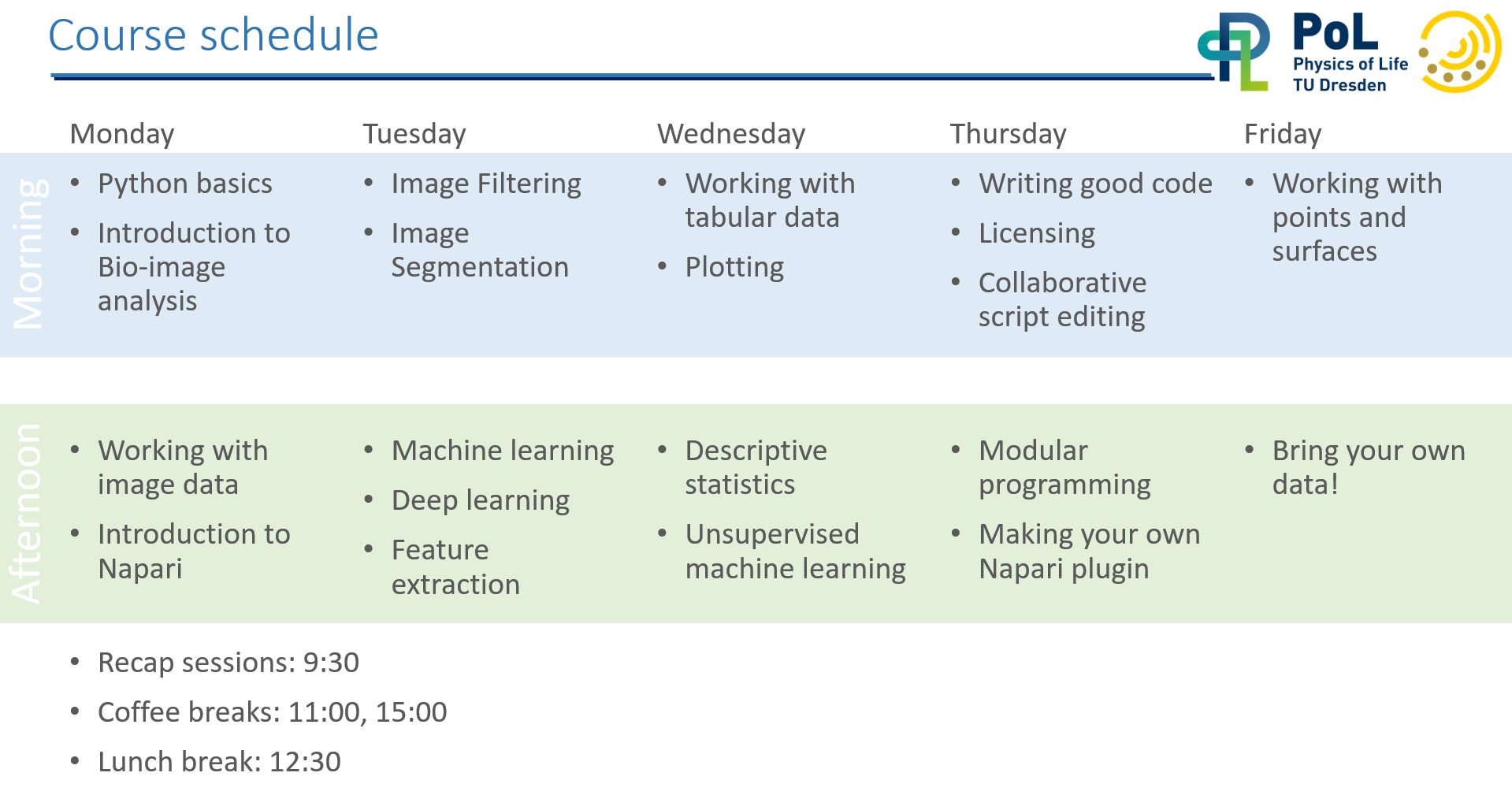
How to use this material#
For following the course, we recommend downloading the repository from which this book is made. All Jupyter Notebooks are executable so that attendees can reproduce all demos and exercises.
Feedback and support#
If you have any questions, please use the anonymous etherpad (see email) or create a github issue. Alternatively, open a thread on image.sc, put a link to the lesson or exercise you want to ask a question about and tag @haesleinhuepf.
Acknowledgements#
We would like to thank all the people who shared teaching materials we are reusing here, in particular Anna Poetsch (Biotec Dresden), Benoit Lombardot (MPI-CBG Dresden), Martin Weigert (EPFL Lausanne) and Alexander Krull (U Birmingham). We acknowledge support by the Deutsche Forschungsgemeinschaft under Germany’s Excellence Strategy—EXC2068–Cluster of Excellence Physics of Life of TU Dresden.
