Removing image noise
Contents
Removing image noise#
In this exercise, we will try out different threshold methods, see how they react to noisy data - and how filtering can help to improve the results. To finish this excercise, have a look at the notebook’s from last week - you may find some useful code!
from skimage import data, filters, measure, util, morphology
import matplotlib.pyplot as plt
import napari
import numpy as np
One thing about variable names#
There are a few conventions for variable names.
No numbers as first letter: Variable names must not begin with a number: A variable called
2nd_variableis invalid in Python - a variable calledvariable2works.Careful when re-using variable names: Above, we imported a module of
skimagethat is calleddata. All functions that are stored in this submodule can be called bydata.some_function(). If you were, to create a variable calleddata(likedata = np.zeros((10,10)), you will overwrite the reference to the module. Because how is Python to know whether, when usingdata, you would like to dodata.some_function()or access your stored values indata?
About pixel noise…#
Pixel noise can have very different characteristics - each of them requires a different type of image filtering to get rid of it. When we talk about noise, we typically differentiate between pixel noise and image noise, both of which have different origins and some subtypes.
Pixel noise: This can, for instance, originate from electronic noise of the camera at the microscope and influences the pixels of the image individually.
# Remember - do not call new variables `data` when you use the skimage module of the same name!
my_data = np.zeros((5,5))
my_data
array([[0., 0., 0., 0., 0.],
[0., 0., 0., 0., 0.],
[0., 0., 0., 0., 0.],
[0., 0., 0., 0., 0.],
[0., 0., 0., 0., 0.]])
There are different types of pixel noise. Gaussian noise, for instance, adds a small value to every pixel according to a gaussian distribution. The gaussian filter provides a good way to deal with this.
data_gaussian_noise = util.random_noise(my_data, mode='gaussian')
data_gaussian_noise
array([[0. , 0.05311451, 0. , 0.08997193, 0. ],
[0. , 0. , 0. , 0.10303216, 0. ],
[0. , 0.01311625, 0.05409787, 0.04064476, 0. ],
[0. , 0.15586629, 0.04207184, 0.07648284, 0.04646911],
[0.08440187, 0.00415252, 0. , 0.13531685, 0.10428321]])
Salt noise changes random pixels to high values - such noise can originate from dead pixels of the detector hardware. The median filter provides a good way to deal with this
data_salt_noise = util.random_noise(my_data, mode='salt')
data_salt_noise
array([[0., 0., 0., 0., 0.],
[0., 0., 0., 0., 0.],
[0., 0., 0., 1., 0.],
[0., 0., 0., 0., 0.],
[0., 0., 0., 0., 1.]])
Conversely, pepper noise can change single pixels to very low values, which can also originate from hardware isues. The median filter also provides a good option to deal with this type of noise.
my_data = np.ones((5,5))
my_data
array([[1., 1., 1., 1., 1.],
[1., 1., 1., 1., 1.],
[1., 1., 1., 1., 1.],
[1., 1., 1., 1., 1.],
[1., 1., 1., 1., 1.]])
data_pepper_noise = util.random_noise(my_data, mode='pepper')
data_pepper_noise
array([[1., 1., 1., 1., 1.],
[1., 1., 1., 1., 1.],
[1., 1., 1., 1., 1.],
[1., 1., 1., 1., 1.],
[1., 0., 1., 1., 1.]])
The image background represents a type of noise that affects large areas of the image, which can orginate from heterogeneous lighting at a microscope or autofluorescence of fluorescent markers. The top-hat filter provides a suitable means to address this problem.
Excercises#
First, we create some data and then artifically add different kinds of noise to it - and see how we can best remove these further down the line again to make our downstream analysis more robust.
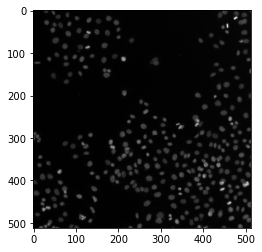
# First, we create some noisy data
image = data.human_mitosis()
plt.imshow(image, cmap='gray')
<matplotlib.image.AxesImage at 0x240a72ce280>
Exercise 1#
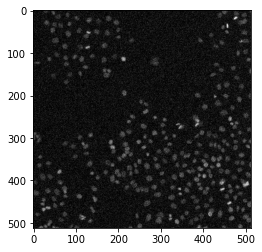
Let’s add some pixel noise to the data. Skimage provides some handy tools for this. Add or remove some of the noise contributions by un-commenting some of the lines below - you can also add multiple sources of noise to the image.
Hint: Use the vmin and vmax parameters to better visualize the brightness and contrast.
noisy_data = util.random_noise(image, mode='gaussian') * 255
#noisy_data = utils.random_noise(image, mode='pepper') * 255
#noisy_data = utils.random_noise(image, mode='s&p') * 255
#noisy_data = utils.random_noise(image, mode='salt') * 255
plt.imshow(noisy_data, cmap='gray', vmin=0, vmax=255)
<matplotlib.image.AxesImage at 0x240a75df0d0>
Exercise 2#
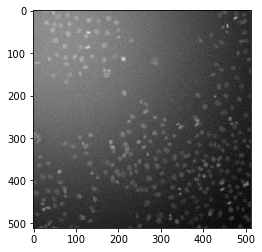
Now, we add another type of noise - background - and add it to the noisy data from above.
Optional: Try to understand the code below. What happens when you modify the value for sigma in line 3?
bg = np.zeros_like(noisy_data)
bg[50,50] = 255
bg_blur = filters.gaussian(bg, sigma=300)
bg_blur = 255*bg_blur/bg_blur.max()
noisy_data += bg_blur
plt.imshow(noisy_data, cmap='gray') # adjust the values for vmin and vmax to get a better impression!
<matplotlib.image.AxesImage at 0x240a7a6bc70>
Exercise 3:#
In order to clear the added noise to the raw image in variable noisy_data, you need to apply noise removal and background subtraction.
Hint 1: The morphology.white_tophat() and filters.gaussian() may provide the correct means to remove some types of noise in the image - but make sure to pass suitable parameters to these functions!
background subtraction with
morphology.white_tophat(): Have a look at the footprint parameter!Noise removal with
filters.gaussian(): Have a look at the sigma parameters!
Hint 2: Look up the documentation for salt & pepper noise - how are pixel values changed if this type of noise is applied? Which filter operation could be suitable to deal with single pixels with high grey values?
background_subtracted =
clean_image =

