PCA (Principle Component analysis)
Contents
PCA (Principle Component analysis)#
Notebook adapted from Anna Poetsch (source) under CC-BY-4.0
Source material:
Tutorial: https://umap-learn.readthedocs.io/en/latest/
Paper: https://arxiv.org/abs/1802.03426
Packages (if not available, pip install):
import numpy as np
from sklearn.preprocessing import StandardScaler
import matplotlib.pyplot as plt
import seaborn as sns
import pandas as pd
Load data:
penguins = pd.read_csv("https://github.com/allisonhorst/palmerpenguins/raw/5b5891f01b52ae26ad8cb9755ec93672f49328a8/data/penguins_size.csv")
penguins = penguins.dropna()

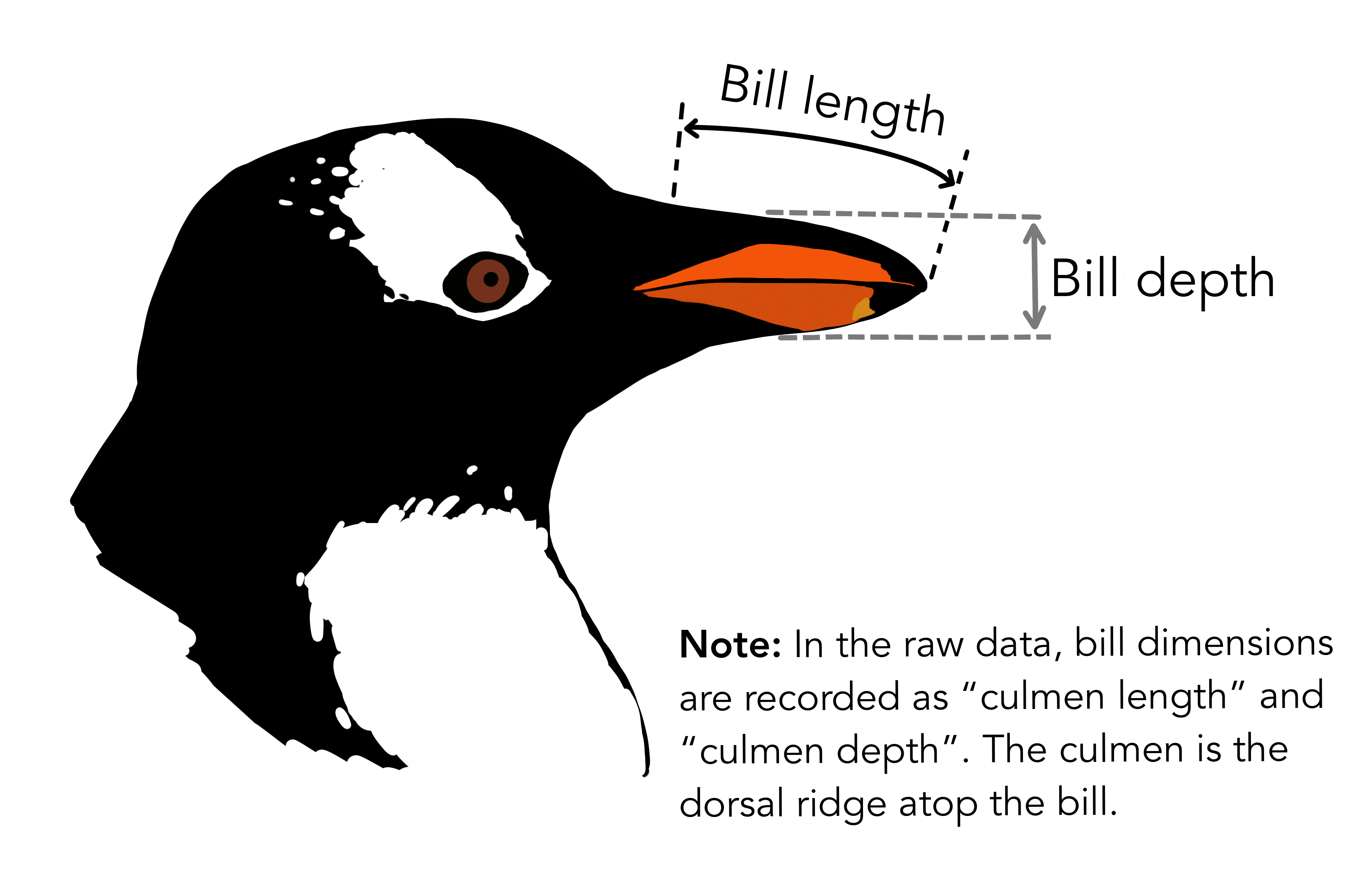
Show data:
penguins.head()
| species_short | island | culmen_length_mm | culmen_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | MALE |
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | FEMALE |
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | FEMALE |
| 4 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | FEMALE |
| 5 | Adelie | Torgersen | 39.3 | 20.6 | 190.0 | 3650.0 | MALE |
penguins = penguins.dropna() penguins.species_short.value_counts()
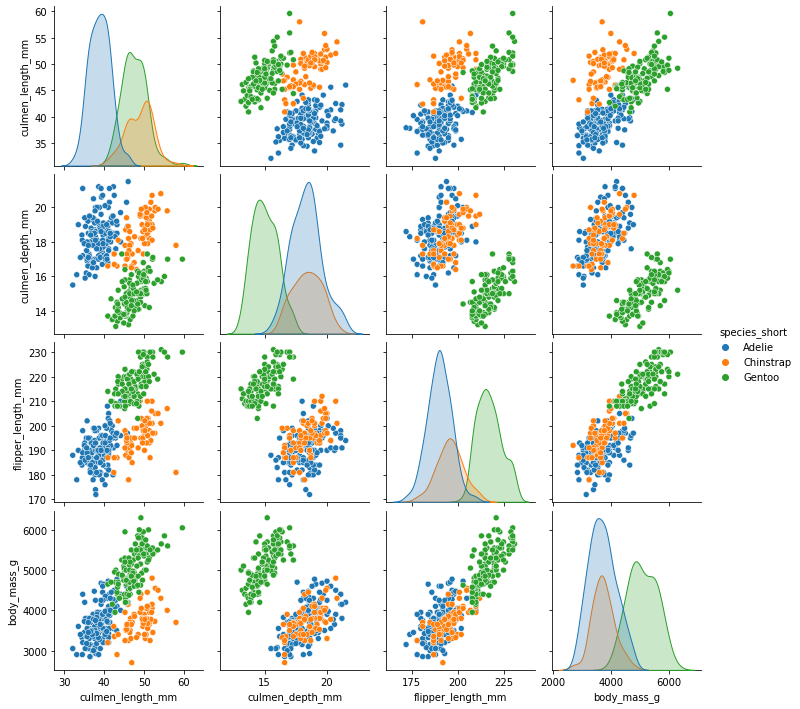
sns.pairplot(penguins, hue='species_short')
<seaborn.axisgrid.PairGrid at 0x7fae38a024f0>
Data scaling#
change data format to values:
penguin_data = penguins[
[
"culmen_length_mm",
"culmen_depth_mm",
"flipper_length_mm",
"body_mass_g",
]
].values
import umap.umap_ as umap #install with 'pip install umap-learn'
np.random.seed(42) #a seed is defined, because there is a random component to UMAP
reducer = umap.UMAP()
scaled_penguin_data = StandardScaler().fit_transform(penguin_data)
scaled_penguin_data
array([[-0.89765322, 0.78348666, -1.42952144, -0.57122888],
[-0.82429023, 0.12189602, -1.07240838, -0.50901123],
[-0.67756427, 0.42724555, -0.42960487, -1.19340546],
...,
[ 1.17485108, -0.74326098, 1.49880565, 1.91747742],
[ 0.22113229, -1.20128527, 0.78457953, 1.23308319],
[ 1.08314735, -0.53969463, 0.85600214, 1.48195382]])
scaled_df = pd.DataFrame(scaled_penguin_data)
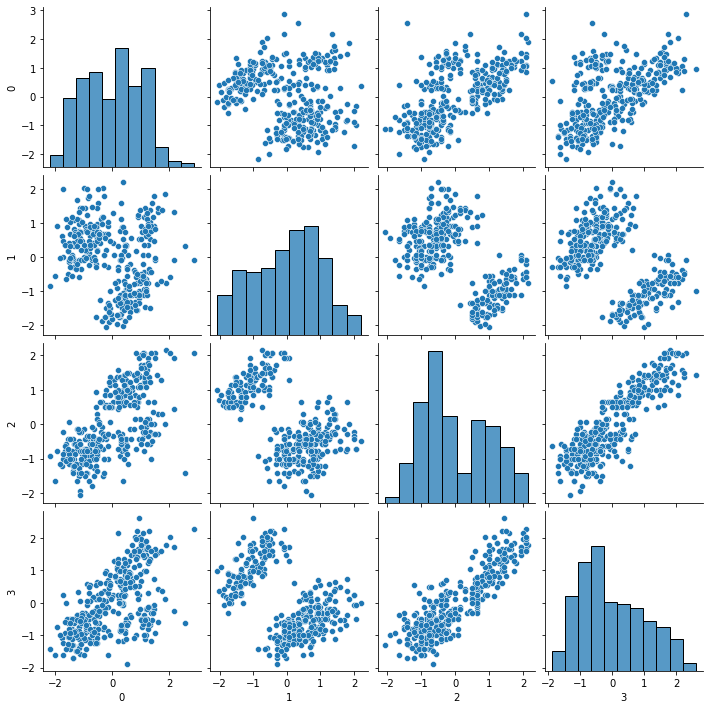
sns.pairplot(scaled_df)
<seaborn.axisgrid.PairGrid at 0x7fae38da70a0>
PCA#
Principle component analysis is a linear dimensionality reduction technique, while UMAP is non-linear. Another popular non-linear one is tSNE.
from sklearn.decomposition import PCA
pca = PCA(n_components=4)
pen_pca = pca.fit(scaled_penguin_data).transform(scaled_penguin_data)
Show the percentage of variance explained for each components:
print(
"explained variance ratio: %s"
% str(pca.explained_variance_ratio_)
)
plt.scatter(
pen_pca[:, 0],
pen_pca[:, 1],
c=[sns.color_palette()[x] for x in penguins.species_short.map({"Adelie":0, "Chinstrap":1, "Gentoo":2})])
plt.show()
explained variance ratio: [0.68641678 0.19448404 0.09215558 0.02694359]
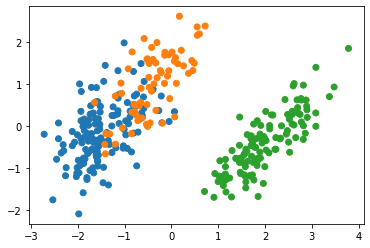
The separation of species with PCA did work, but not very well.

