Executing clesperanto on the TU Dresden HPC#
To execute Python Jupyter notebooks on TU Dresden’s HPC cluster, navigate to this URL: https://taurus.hrsk.tu-dresden.de/jupyter/hub/home
Note: If you’re outside the university network, you need to connect via VPN.
After loggin in, a Jupyter Hub login screen will open. Click on Start My Server.
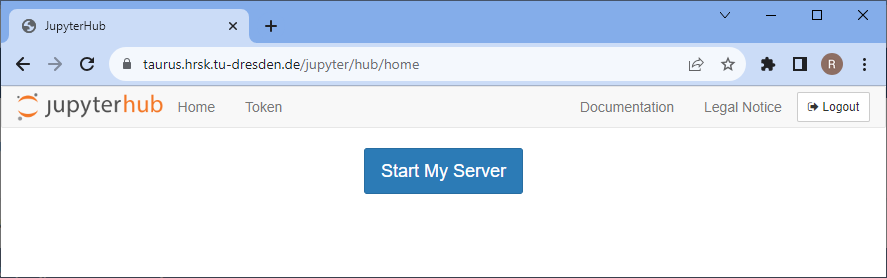
Select the entries as shown below in the Advanced tab and click on Spawn.
This will take a moment.
You will be redirected to a Jupyter Lab environment.
Now you need to activate the install a singularity container on the HPC cluster as explained in detail here. In short:
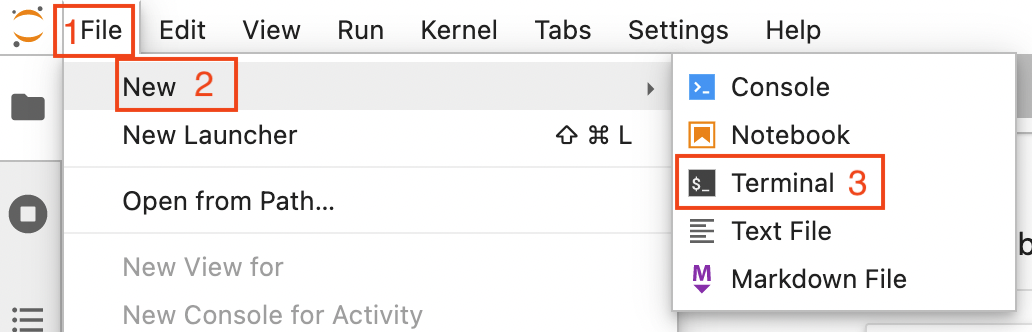
Open a terminal by clicking on File (1 in the image below) -> New (2) -> Terminal (3)
Install a custom jupyter kernel for your user
To install a devbio-napari python environment, execute the following code in the terminal:
git clone https://gitlab.mn.tu-dresden.de/bia-pol/singularity-devbio-napari.git
cd singularity-devbio-napari
./install.sh v0.2.9
Wait 2-15 min until the image is downloaded and verified (the time depends on how much network and disk load is on the cluster). The output should look something like this:

Now reload the browser tab.
Use the upload button to upload your assistant-generated notebook. You can also use this example notebook.
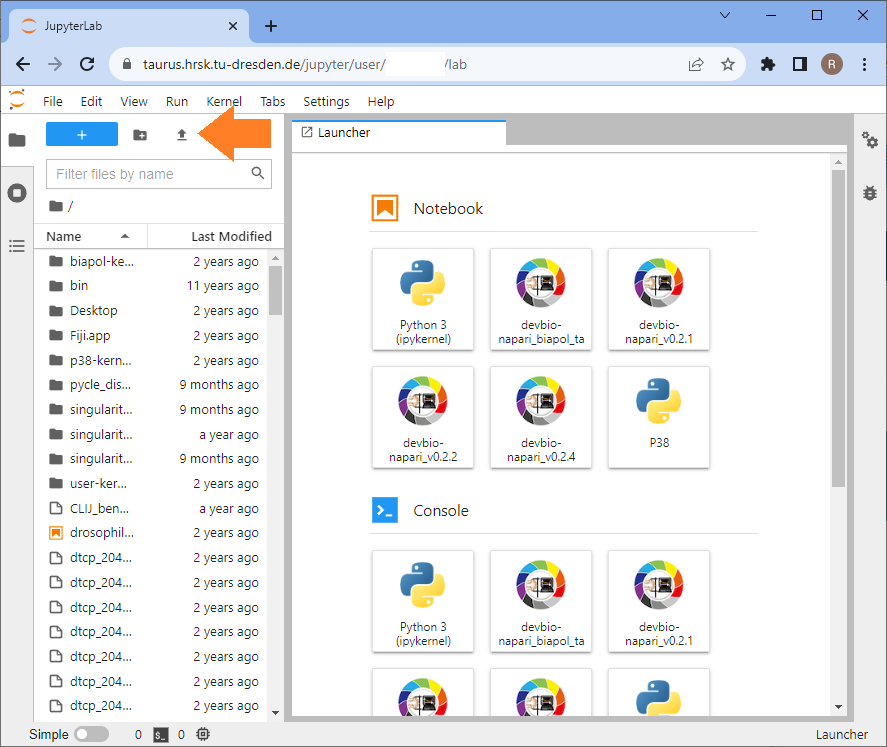
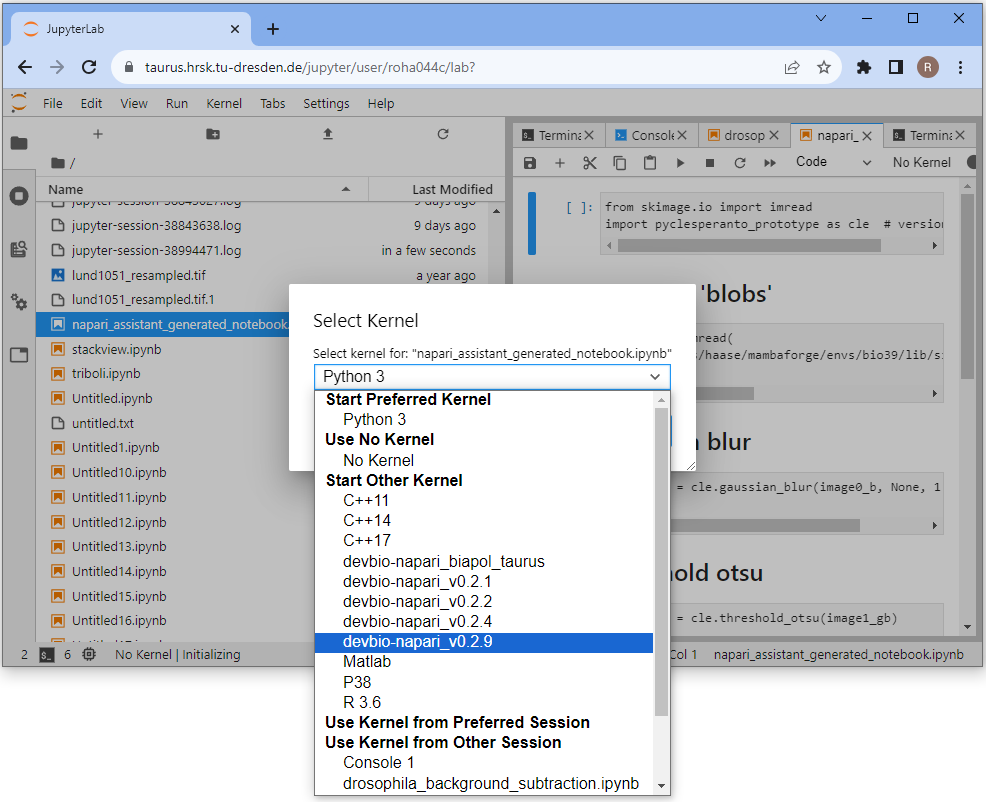
Double-click the uploaded notebook to open it. Select a kernel, e.g. the devbio-napari kernel.
In this notebook, you need to change how files are opened.
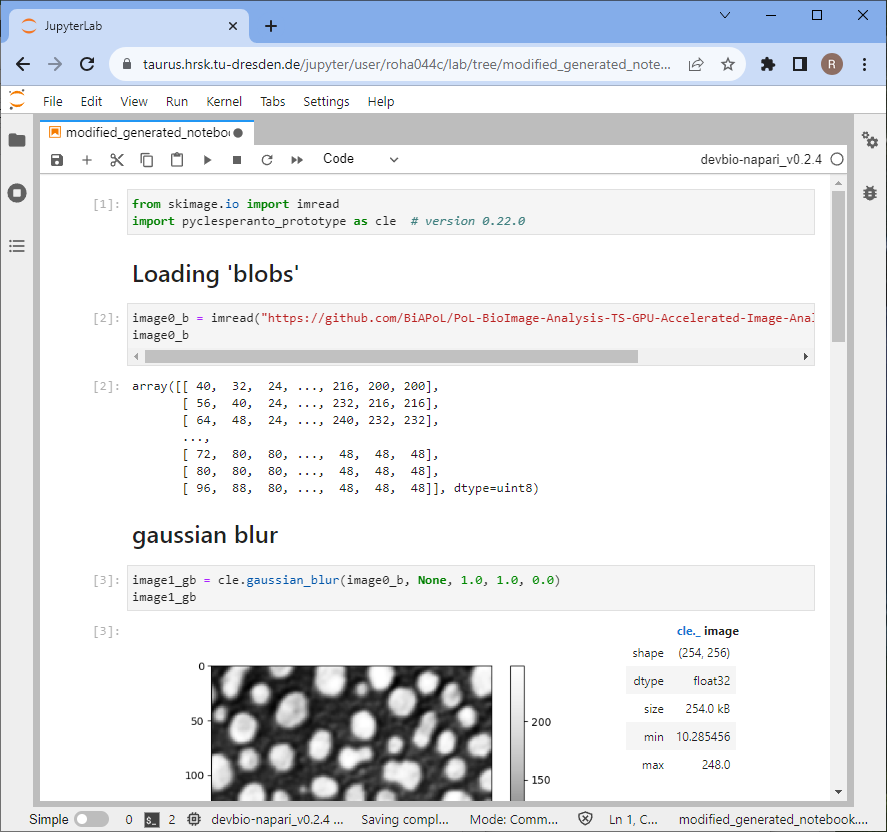
You find a potential modification on the following page.
