Edge handling experiments#
In this example we look closer at edge handling, this time in 3D.
For background on the derivation of the Richardson Lucy equation for non-circulant boundaries see this Application note
When image is extended we can think of it being equivalent to extending our ‘solution space’. Then the acquired image has been multiplied by a window w in the larger space. And the RL equation becomes.
Where n is a normalization factor calculated as HTones. The H operator on the image is convolution followed by multiplication by a window
So HTones would be window applied to the ones matrix followed by convolution by the conjugate of the PSF.
Load the test image#
Load the test image and crop the image a bit as to speed up testing. We also reverse the z axis of the stack so that it is consistent with positive ‘depth’ being below the coverslip. We have cropped the test image to better demonstrate the performance of different edge handling approaches the cropped version of the image can be found here and the original image can be found here
from skimage.io import imread
import numpy as np
from tnia.plotting.projections import show_xy_zy_slice
from decon_helper import image_path
bead=imread(image_path / "bead-2.5um.tif")
z_to_view=bead.shape[0]//2
xy_spacing = 0.0645
z_spacing=.16
ratio = z_spacing/xy_spacing
print(bead.shape)
fig=show_xy_zy_slice(bead,128,128,z_to_view, sz=ratio, figsize=(7,4))
(50, 256, 256)
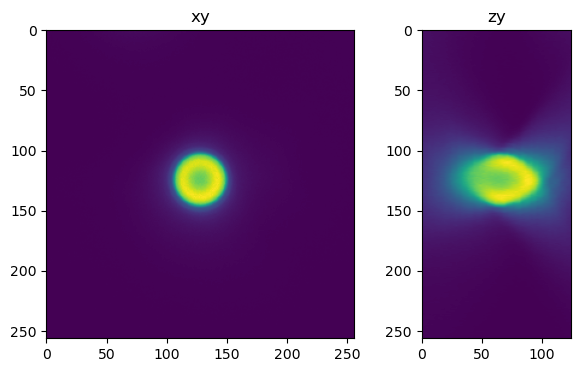
Create the PSF#
Here we generate the PSF. The meta data does not include the refractive index of the medium the bead is embedded in. It is likely close to, but not exactly the refractive index of the lens immersion media, in a previous experiment we dtermined the RI of the embedding media to be apr. 1.49.
from tnia.nd.ndutil import centercrop
from tnia.deconvolution.psfs import gibson_lanni_3D
from tnia.plotting.projections import show_xy_zy_max
from tnia.deconvolution.psfs import recenter_psf_axial
from skimage.io import imsave
ni=1.518
ns = 1.49
NA=1.4
w=0.530
xy_spacing = 0.0645
z_spacing=.16
xy_psf_dim=255
z_compute_psf_dim=101
#z_crop_psf_dim=101
#depth to compute PSF at
d=128*z_spacing
psf = gibson_lanni_3D(NA, ni, ns, xy_spacing, z_spacing, xy_psf_dim, z_compute_psf_dim, d, w, False, True)
psf = psf.astype('float32')
psf = psf/psf.sum()
#psf=recenter_psf_axial(psf, 255)
fig=show_xy_zy_max(psf, vmax=psf.max()/4, figsize=(8,4))
Set deconvolution parameters#
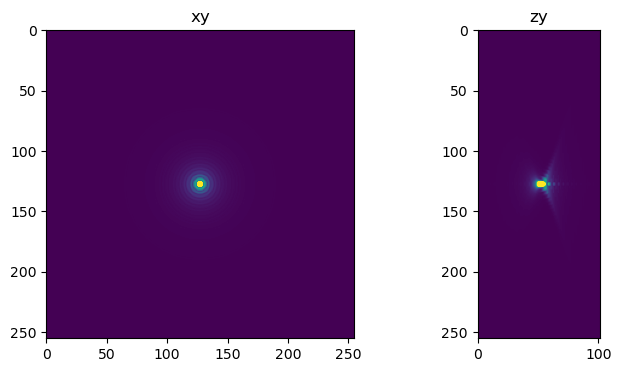
For the first experiment we deconvolve a centered bead for 2000 iterations. Even though the bead is centered we still see artifacts when proper edge handling is not used.
from clij2fft.richardson_lucy import richardson_lucy_nc, richardson_lucy
import RedLionfishDeconv as rl
bead = bead.astype('float32')
regularization_factor=0
iterations=1000
Perform deconvolution with RedLionFish, CLIJ Non-circulant, and CLiJ Circulant#
import RedLionfishDeconv as rl
decon_rl=rl.doRLDeconvolutionFromNpArrays(bead, psf, niter=iterations, method='gpu', resAsUint8=False )
regularization_factor=0
im_decon_clij_nc = richardson_lucy_nc(bead, psf, iterations , regularization_factor)
im_decon_clij= richardson_lucy(bead, psf, iterations , regularization_factor)
get lib
get lib
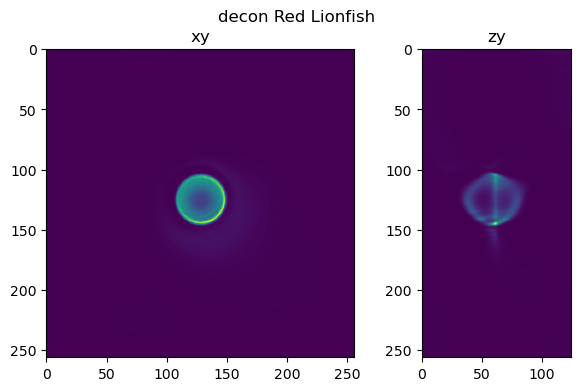
fig=show_xy_zy_slice(decon_rl,128,128,z_to_view, sz=ratio, figsize=(7,4))
fig.suptitle('decon Red Lionfish')
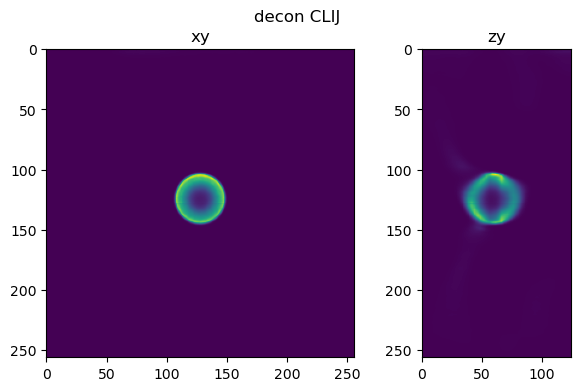
fig=show_xy_zy_slice(im_decon_clij,128,128,z_to_view, sz=ratio, figsize=(7,4))
fig.suptitle('decon CLIJ')
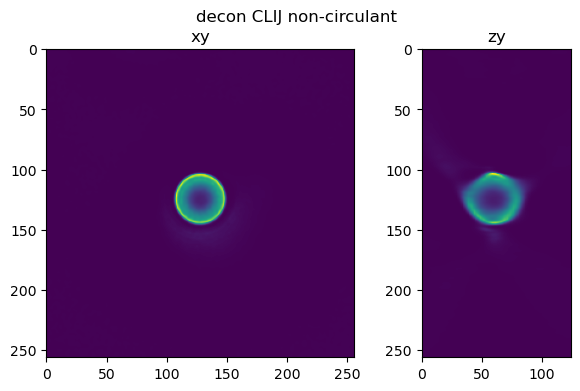
fig=show_xy_zy_slice(im_decon_clij_nc,128,128,z_to_view, sz=ratio, figsize=(7,4))
fig.suptitle('decon CLIJ non-circulant')
Text(0.5, 0.98, 'decon CLIJ non-circulant')
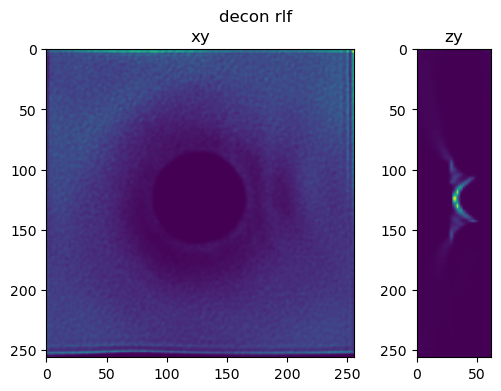
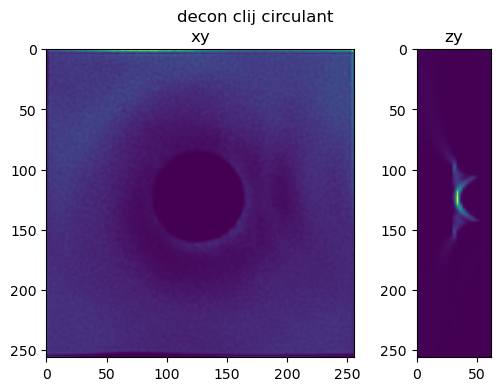
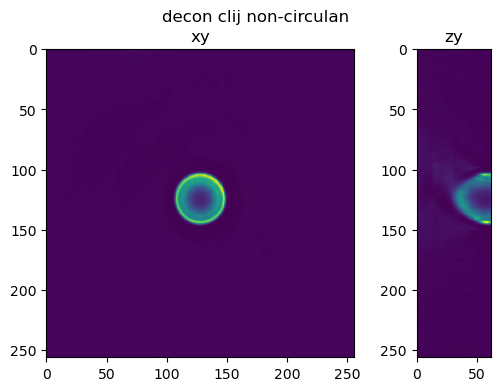
Perform the same tests using a half bead#
half_bead=bead[:bead.shape[0]//2,:,:]
z_to_view=half_bead.shape[0]-1
fig=show_xy_zy_slice(half_bead,128,128,z_to_view, sz=ratio, figsize=(6,4))
fig.suptitle('half bead')
Text(0.5, 0.98, 'half bead')
psf_crop=centercrop(psf, (half_bead.shape[0], psf.shape[1], psf.shape[2]))
decon_rl=rl.doRLDeconvolutionFromNpArrays(half_bead, psf_crop, niter=iterations, method='gpu', resAsUint8=False )
im_decon_clij_nc = richardson_lucy_nc(half_bead, psf_crop, iterations , regularization_factor)
im_decon_clij = richardson_lucy(half_bead, psf_crop, iterations , regularization_factor)
get lib
get lib
fig=show_xy_zy_slice(half_bead,128,128,z_to_view, sz=ratio, figsize=(6,4))
fig.suptitle('half bead')
fig=show_xy_zy_slice(decon_rl,128,128,z_to_view, sz=ratio, figsize=(6,4))
fig.suptitle('decon rlf')
fig=show_xy_zy_slice(im_decon_clij,128,128,z_to_view, sz=ratio, figsize=(6,4))
fig.suptitle('decon clij circulant')
fig=show_xy_zy_slice(im_decon_clij_nc,128,128,z_to_view, sz=ratio, figsize=(6,4))
fig.suptitle('decon clij non-circulant')
Text(0.5, 0.98, 'decon clij non-circulan')