Exercise: Seeded watershed in Napari
Contents
Exercise: Seeded watershed in Napari#
Start Napari and open the example image File > Open Samples > Napari > Cells (3D+2Ch).
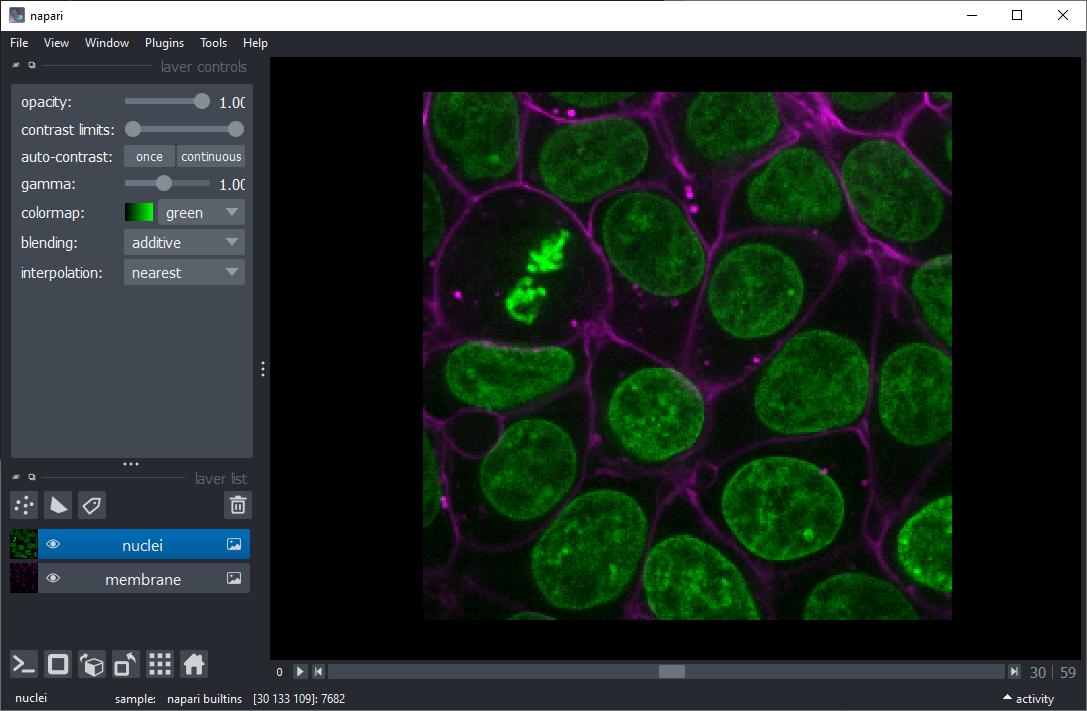
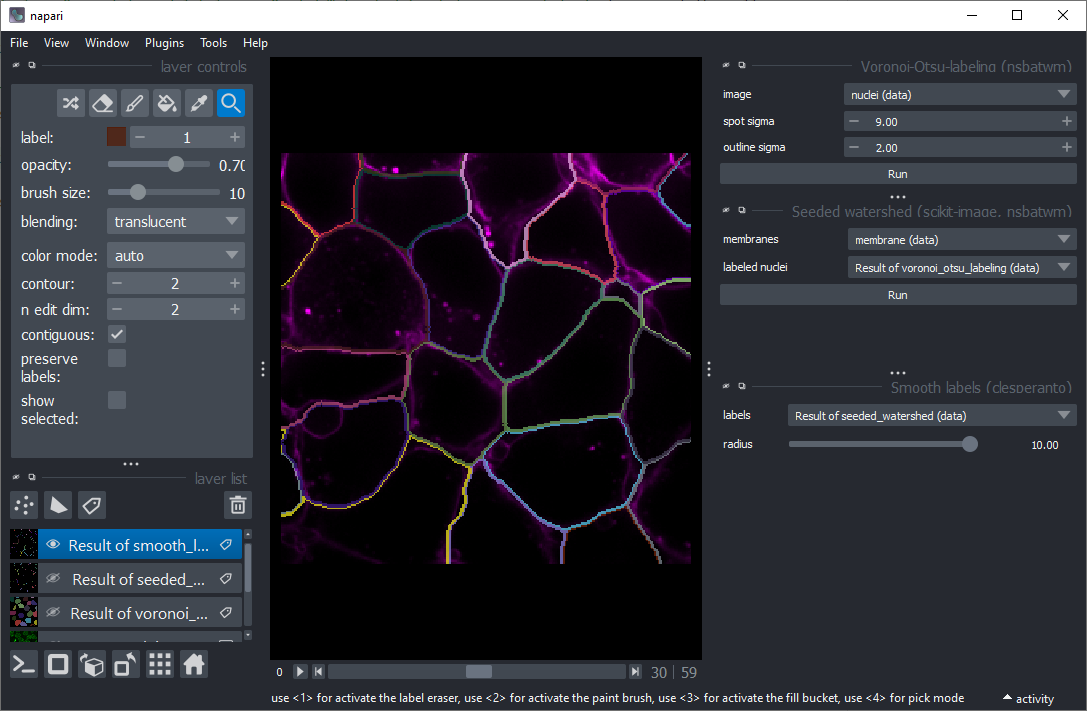
Use the menu Tools > Segmentation / labeling > Voronoi Otsu Labeling to segment the nuclei.
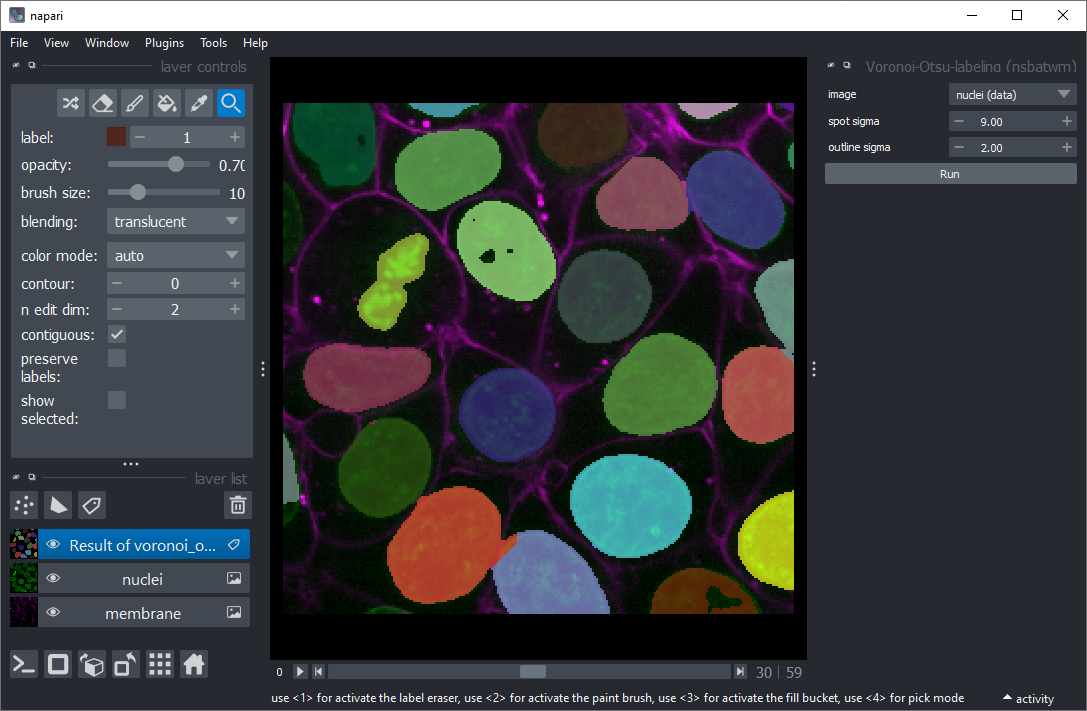
Use the menu Tools > Segmentation / labeling > Seeded Watershed (scikit-image, nsbatwm) to segment the cells.
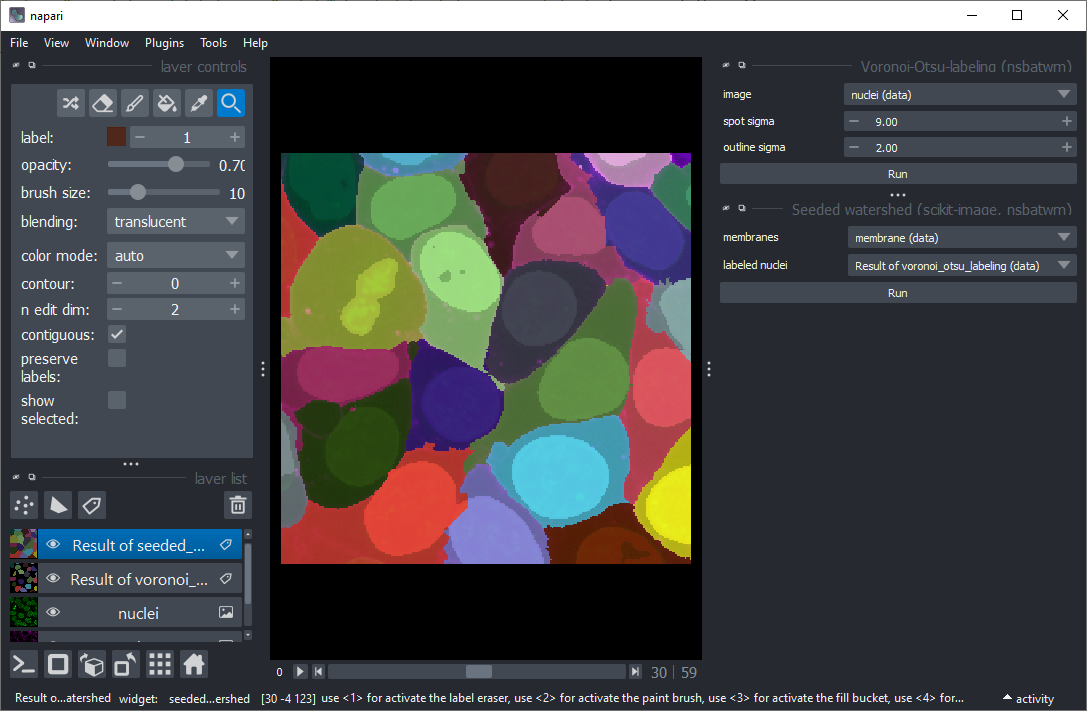
Start Smooth labels (clesperanto) from the menu Tools > Segmentation post-processing.
Hide all layers except the final cell segmentation. Change the contour of the cell segmentation layer to show the outlines of the segmented cells above the membrane channel.
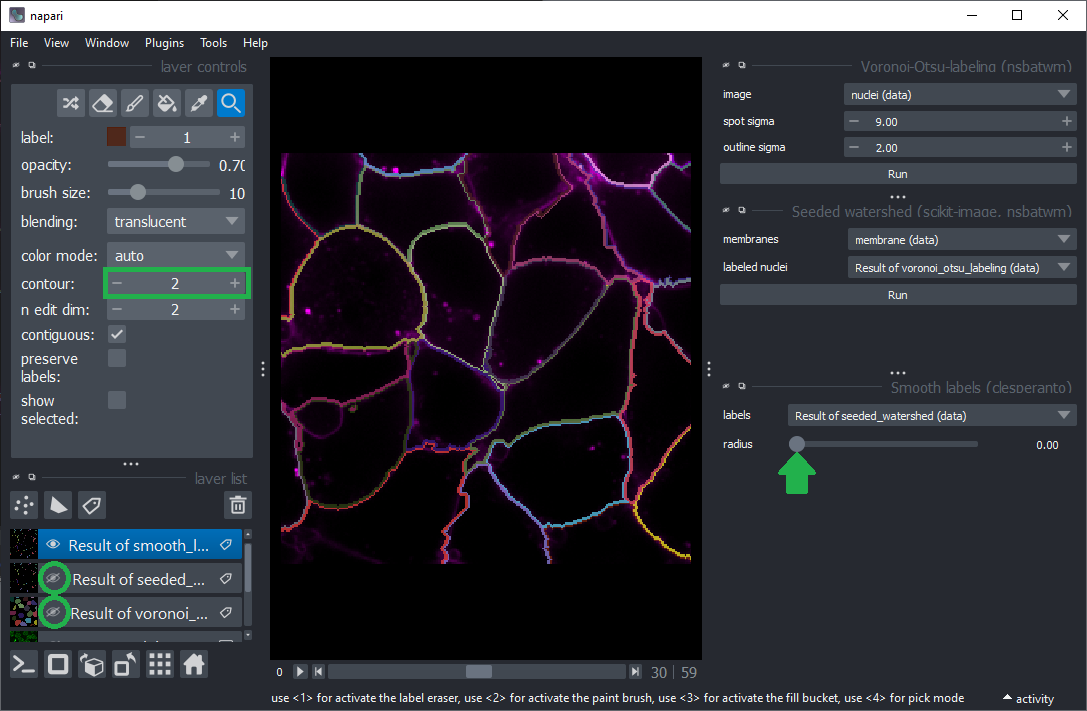
Vary the radius and see how the outlines change.
Question#
What is a good radius for segmenting cells in this image properly?
