Sphericity & Solidity#
In this notebook you will learn how to calculate two common 3D shape descriptors for assessing how similar an object is to a perfect sphere. You will also compare object volume measurements from an image and its corresponding mesh representation.
Please download the required file for the variable binary_gastruloid from this link and add it to your data folder for this session:
https://drive.google.com/file/d/1zywDJeNinfoJC49qFuskqxQ4HGniGSGK/view?usp=sharing
You should not need a password. You will need to have napari-process-points-and-surfaces and the vedo library installed. They should already be part of your devbio-napari-env. If they are not, you can add them using mamba install napari-process-points-and-surfaces and mamba install vedo
The data used in this notebook is derived from AV Luque and JV Veenvliet (2023) licensed CC-BY and can be downloaded from here: https://zenodo.org/record/7603081. Compared to the original, the file used here is binary to reduce file size.
import napari
import napari_process_points_and_surfaces as nppas
import numpy as np
import vedo
from skimage.io import imread, imshow
from skimage.measure import regionprops
import matplotlib.pyplot as plt
%matplotlib inline
Load Image Data#
binary_gastruloid = imread('data/binary_gastruloid.tif')
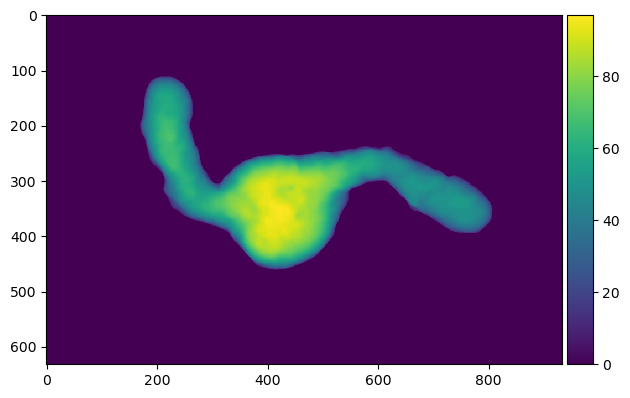
imshow(binary_gastruloid.sum(axis = 0)) # sum projection visualization of binary 3d object
/Users/ryan/mambaforge/envs/devbio-napari-env/lib/python3.9/site-packages/skimage/io/_plugins/matplotlib_plugin.py:150: UserWarning: Low image data range; displaying image with stretched contrast.
lo, hi, cmap = _get_display_range(image)
<matplotlib.image.AxesImage at 0x1b42d5190>
Load Mesh Data#
Use vedo which was introduced to you this morning to load a previously created mesh representation of the above gastruloid imaging dataset.
mesh_gastruloid = vedo.load('data/gastruloid.ply')
Visualize Mesh#
Use the napari_process_points_and_surfaces module to convert the vedo mesh into a SurfaceTuple that can be pushed to a napari viewer as a surface layer.
napari_mesh_gastruloid = nppas.to_napari_surface_data(mesh_gastruloid)
napari_mesh_gastruloid
|
|
nppas.SurfaceTuple
|
Push your image & your mesh to a napari viewer#
Briefly have a look at the overlay of your image and surface layers in napari. It is best to use the 3d view so that you can spin the surface around and zoom in :)
viewer = napari.Viewer()
viewer.add_image(binary_gastruloid)
viewer.add_surface(napari_mesh_gastruloid)
WARNING: Window position QRect(0,0 640x480) outside any known screen, using primary screen
<Surface layer 'napari_mesh_gastruloid' at 0x1b4354e20>
WARNING: Window position QRect(592,-550 100x30) outside any known screen, using primary screen
Extract Mesh Volume & Surface Area#
Here we take advantage of the many measurements that are immediately accessible from vedo.Mesh. For our downstream calculatin of sphericity, let’s extract the mesh’s volume and surface area.
mesh_volume = mesh_gastruloid.volume()
mesh_volume
4690998.704897018
mesh_surface_area = mesh_gastruloid.area()
mesh_surface_area
229569.5362677695
Image Volume#
Let’s also calculate the volume of the binary gastruloid in image space (i.e., discretised space) by taking the count of nonzero voxels.
If you were working with a dataset with several objects, you could choose to apply skiamge.measure.regionprops and then retrive the equivalent values by using the key area. However, because this image only has one object, it is simple enough to use numpy.sum.
binary_volume = np.sum(binary_gastruloid)
binary_volume
4712181
Calculate Sphericity#
Here I have written a function to calculate sphericity.
Sphericity is a very common metric for biological object shape assessment and is simply a specific ratio of volume and surface area. As you can see, the function takes two inputs: a volume measurement and a surface area measurement.
There are plenty of packages in python with functions to calculate sphericity; however, it is important to know (and understand) how they have defined the measurement. The one I have written here is probably the most common mathematical definiton. It has a range of (0,1) and is scalar (i.e., dimensionless). Some implementations you may find could have a different range depending on how the ratio is defined.
def sphericity(mesh_volume, mesh_surface_area):
'''This definition of sphericity comes from a formula that assumes your
object exists in continuous space.
Parameters:
-----------
mesh_volume: integer or float value
mesh_surface_area: integer or float value
Returns:
--------
psi: a float value with range(0,1) reflecting the compactness of an object
'''
from numpy import pi
numerator = (pi ** (1/3)) * ((6 * mesh_volume) ** (2/3))
denominator = mesh_surface_area
psi = numerator / denominator
return psi
sphericity = sphericity(mesh_volume, mesh_surface_area)
sphericity
0.5903099128683961
Convex Hull#
Say you want to compare the sphericity measurement to another metric that should be correlated, but not identical. The example we will use here is solidity, which is the ratio of an object’s volume to the volume of its convex hull. Therefore we must first create the convex hull of our mesh and then measure its volume.
convex_hull_gastruloid = nppas.create_convex_hull_from_surface(napari_mesh_gastruloid)
convex_hull_gastruloid
|
|
nppas.SurfaceTuple
|
convex_hull = nppas.to_vedo_mesh(convex_hull_gastruloid)
convex_hull_volume = convex_hull.volume()
convex_hull_volume
8379992.083160304
Now that we have the convex hull volume we can calculate the mesh’s solidity.
gastruloid_solidity = mesh_volume / convex_hull_volume
gastruloid_solidity
0.5597855771634482
Exercise#
Calculate the sphericity of the gastruloid using image_volume instead of mesh_volume then find the percent difference between the two measurements. Is it statistically significant?
Exercise#
Apply regionprops from skimage.measure to binary_gastruloid and index the resultant dictionary to return the function’s solidity measure. Is it identical (or statistically close enough) to what you computed above?
