Sample data#
The napari-clusters-plotter comes with a number of sample data files for you to test and play with the functionality of the plugin. Here we list where these sample data have been taken from and how to load them into napari.
Labels: BBBC1-Data#
For demo raw image and segmentations as label images, we used image set BBBC007v1 image set version 1 (Jones et al., Proc. ICCV Workshop on Computer Vision for Biomedical Image Applications, 2005), available from the Broad Bioimage Benchmark Collection [Ljosa et al., Nature Methods, 2012]. Images were cropped with a 340x340 window size and converted to 16-bit format. The segmentations have been created using the voronoi-otsu-algorithm from napari-segment-things-with-blobs-and-membranes. The dataset includes several images and their corresponding segmentations side-by-side in the napari viewer.
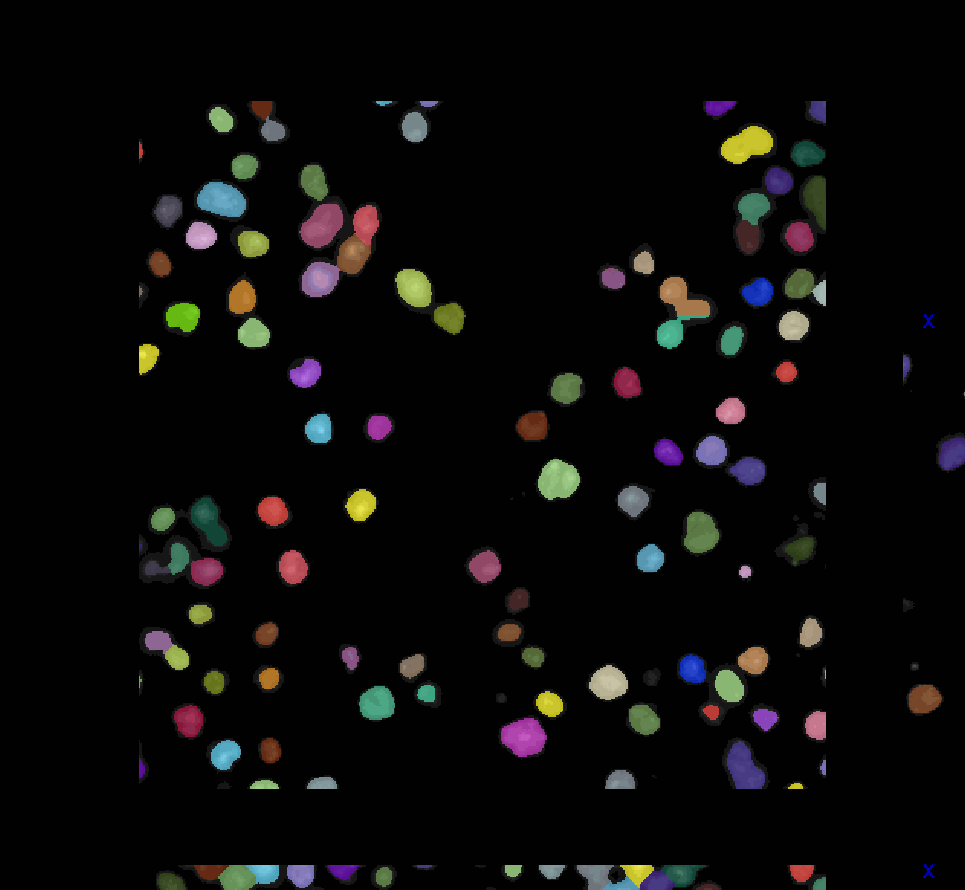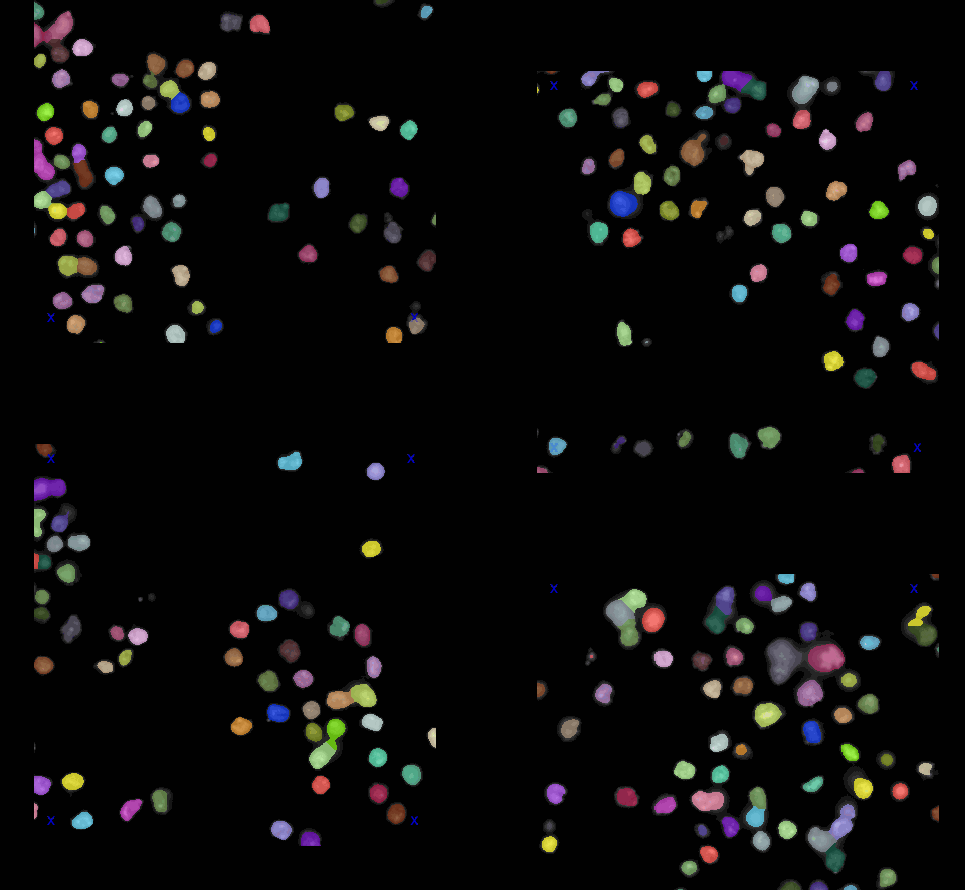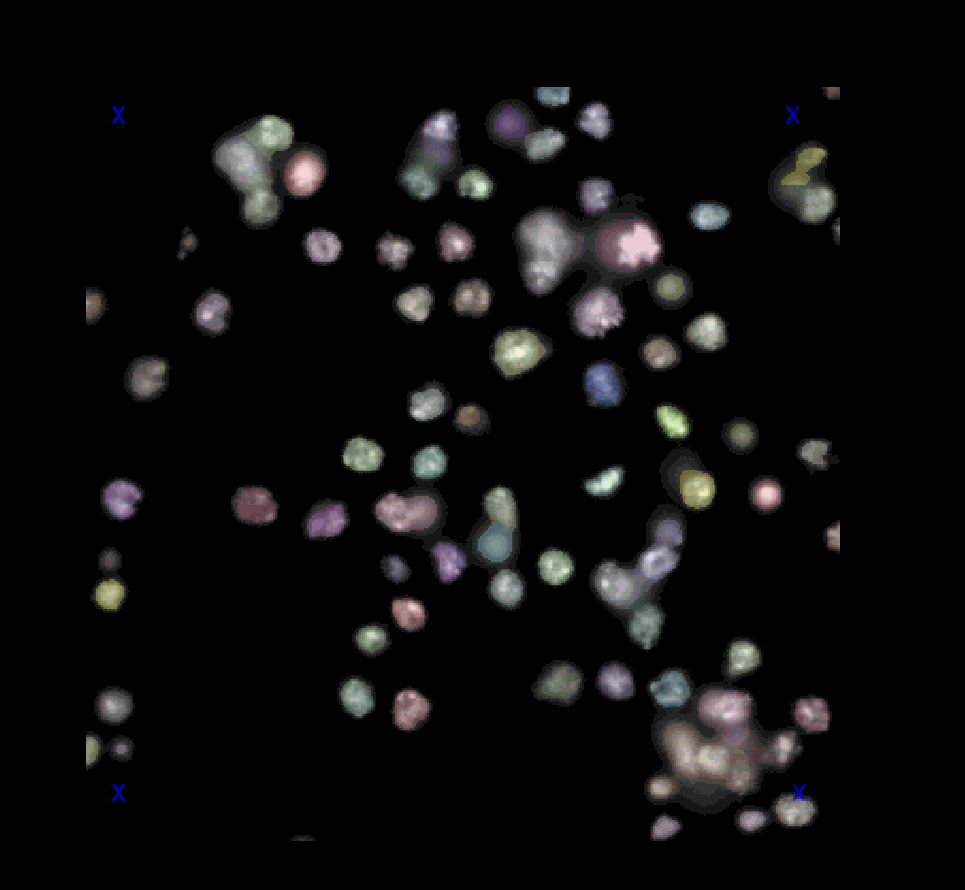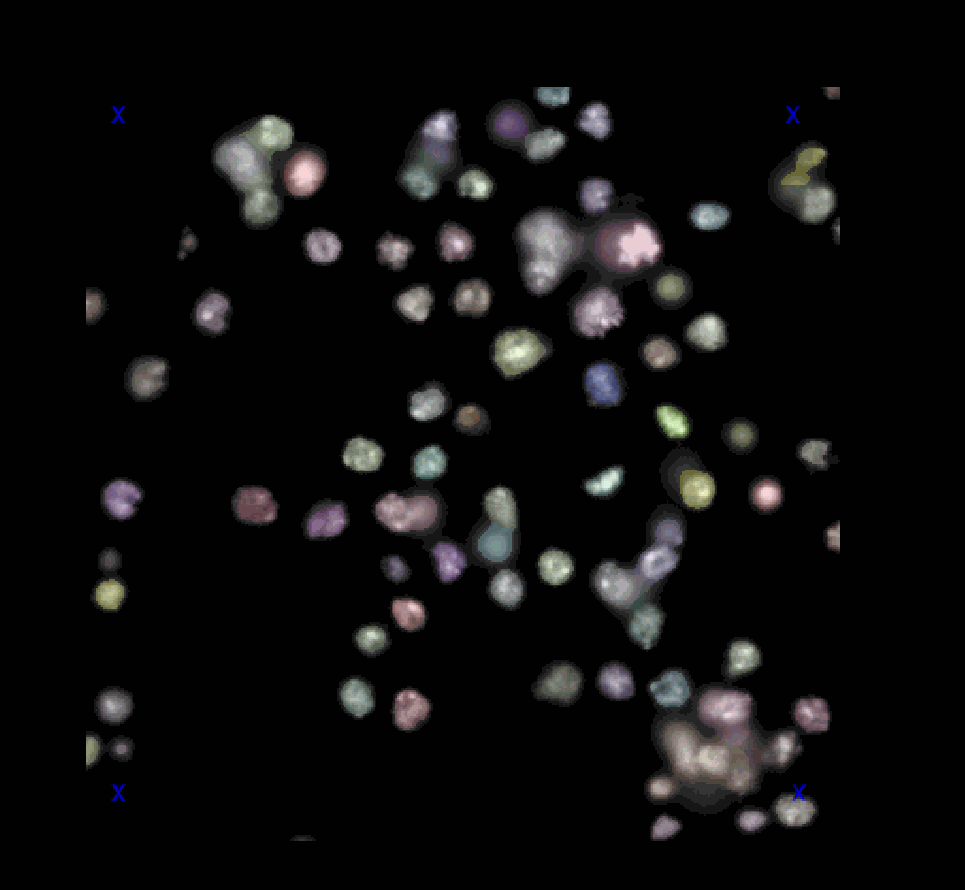
The dataset is available in the napari viewer under File > Open Sample > napari clusters plotter > BBBC 1 dataset & segmentations
Surface: Cells3d curvatures#
For surface (aka mesh) data, we segmented and converted a piece of the scikit-image cells3d example dataset. The data were converted according to this notebook from the napari-process-points-and-surfaces plugin. The dataset includes a raw image of a mitotic cell and its corresponding surface mesh. Mean and Gaussian curvatures have been measured on the surface with the patch-fitting algorithm taken from the napari-stress plugin
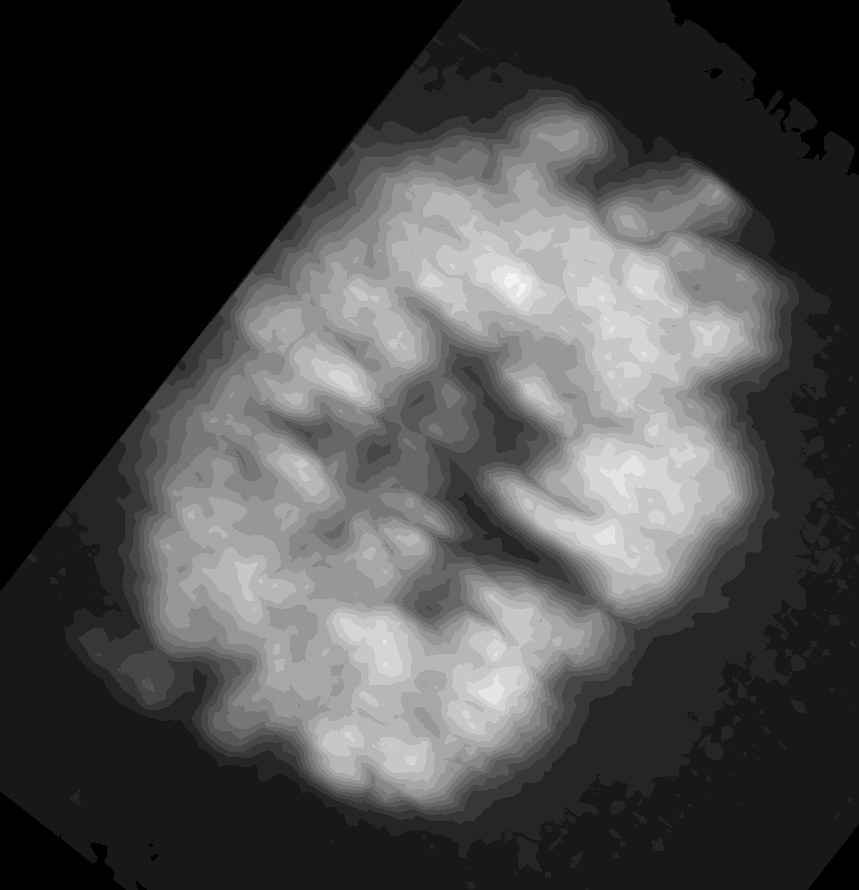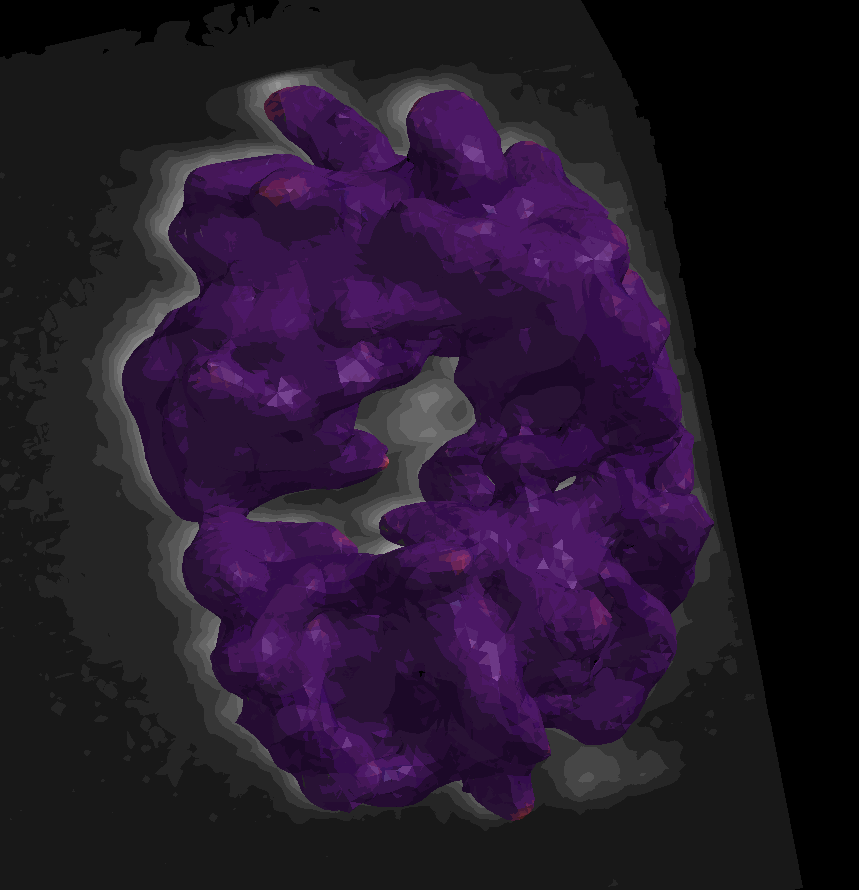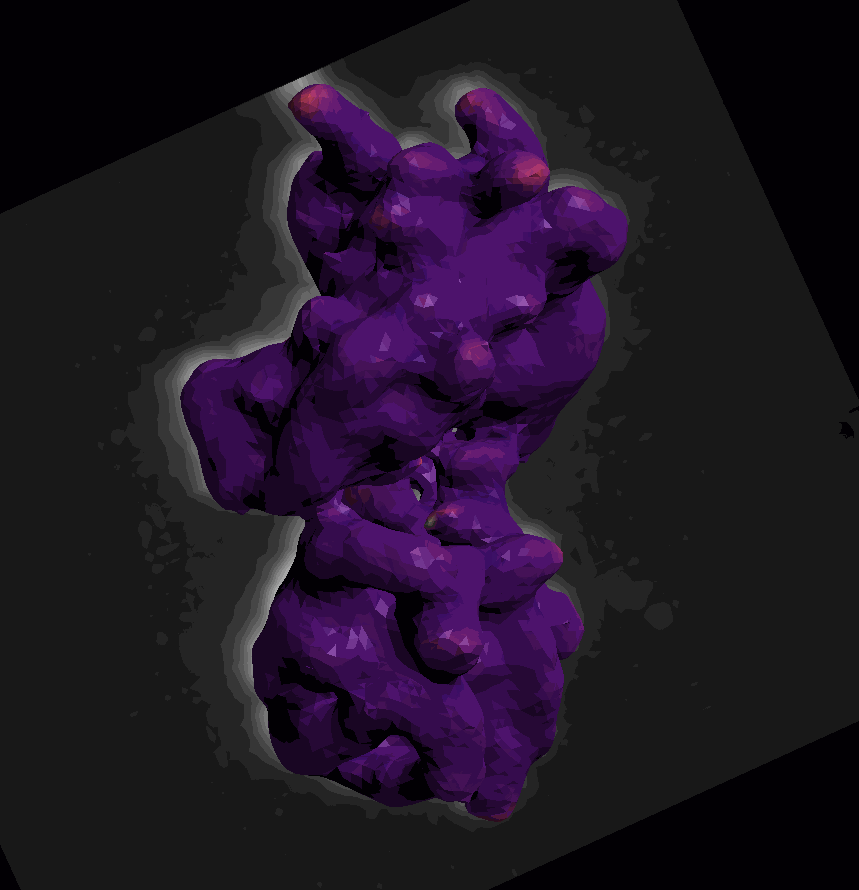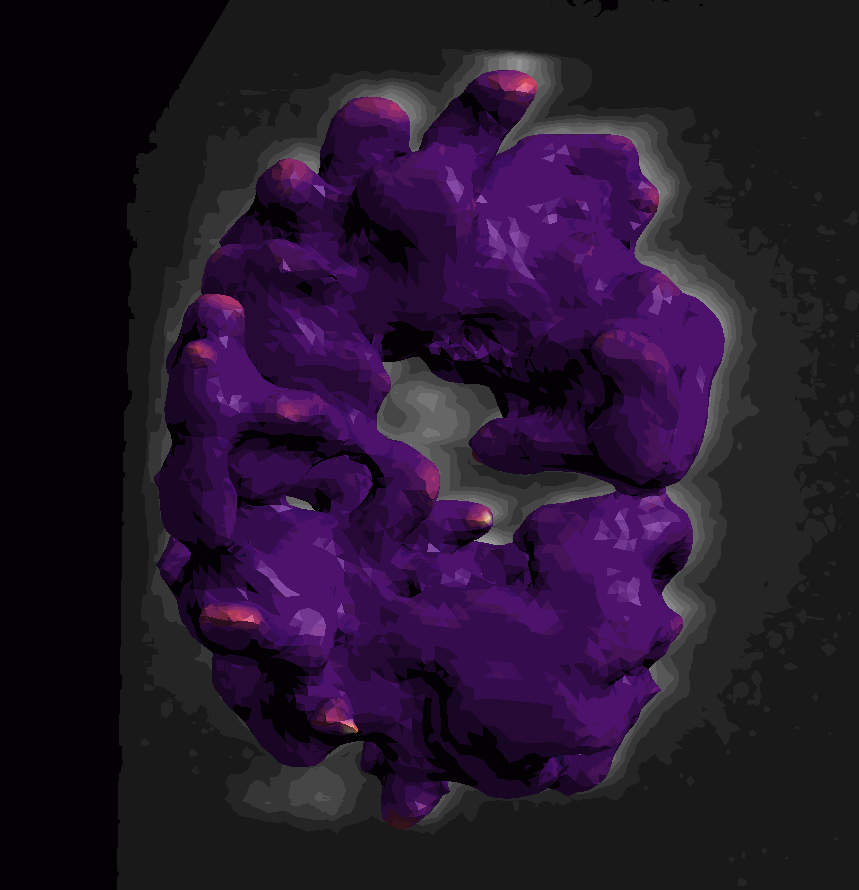
The dataset is available in the napari viewer under File > Open Sample > napari clusters plotter > Cells3D mitotic nucleus surface curvatures
Tracks: tgmm-mini labels and tracks#
For tracking data, we provide the tgmm-mini dataset which includes segmented nuclei that are tracked over time and label-matched so that nuclei retain their label over time. It has been derived from tgmm-mini using Fiji with the Mastodon and the Mastodon-Deep-Lineage plugin.
The data is licensed under the conditions in LICENSE.
The dataset is available in the napari viewer under File > Open Sample > napari clusters plotter > TGMM mini dataset (tracks and segmentations)
Shapes: Skan skeleton#
For shapes data, we provide a set of skeleton paths that were determined from a random dataset using the skan library according to this workflow.

The dataset is available in the napari viewer under File > Open Sample > napari clusters plotter > Skan skeleton dataset(labels and paths)
